Computes a tree model with networks at the end of branches. Can use model-based recursive partitioning or conditional inference.
Wraps the mob() and ctree() functions from the partykit package.
networktree(...)
# S3 method for default
networktree(
nodevars,
splitvars,
method = c("mob", "ctree"),
model = "correlation",
transform = c("cor", "pcor", "glasso"),
na.action = na.omit,
weights = NULL,
...
)
# S3 method for formula
networktree(
formula,
data,
transform = c("cor", "pcor", "glasso"),
method = c("mob", "ctree"),
na.action = na.omit,
model = "correlation",
...
)Arguments
- ...
additional arguments passed to
mob_control(mob) orctree_control(ctree)- nodevars
the variables with which to compute the network. Can be vector, matrix, or dataframe
- splitvars
the variables with which to test split the network. Can be vector, matrix, or dataframe
- method
"mob" or "ctree"
- model
can be any combination of c("correlation", "mean", "variance") splits are determined based on the specified characteristics
- transform
should stored correlation matrices be transformed to partial correlations or a graphical lasso for plotting? Can be set to "cor" (default), "pcor", or "glasso"
- na.action
a function which indicates what should happen when the data contain missing values (
NAs).- weights
weights
- formula
A symbolic description of the model to be fit. This should either be of type
y1 + y2 + y3 ~ x1 + x2with node vectorsy1,y2, andy3ory ~ x1 + x2with a matrix response y.x1andx2are used as partitioning variables.- data
a data frame containing the variables in the model
References
Jones, P.J., Mair, P., Simon, T., Zeileis, A. (2020). Network trees: A method for recursively partitioning covariance structures. Psychometrika, 85(4), 926-945. https://doi.org/10.1007/s11336-020-09731-4
Examples
set.seed(1)
d <- data.frame(trend = 1:200, foo = runif(200, -1, 1))
d <- cbind(d, rbind(
mvtnorm::rmvnorm(100, mean = c(0, 0, 0),
sigma = matrix(c(1, 0.5, 0.5, 0.5, 1, 0.5, 0.5, 0.5, 1), ncol = 3)),
mvtnorm::rmvnorm(100, mean = c(0, 0, 0),
sigma = matrix(c(1, 0, 0.5, 0, 1, 0.5, 0.5, 0.5, 1), ncol = 3))
))
colnames(d)[3:5] <- paste0("y", 1:3)
## Now use the function
tree1 <- networktree(nodevars=d[,3:5], splitvars=d[,1:2])
## Formula interface
tree2 <- networktree(y1 + y2 + y3 ~ trend + foo, data=d)
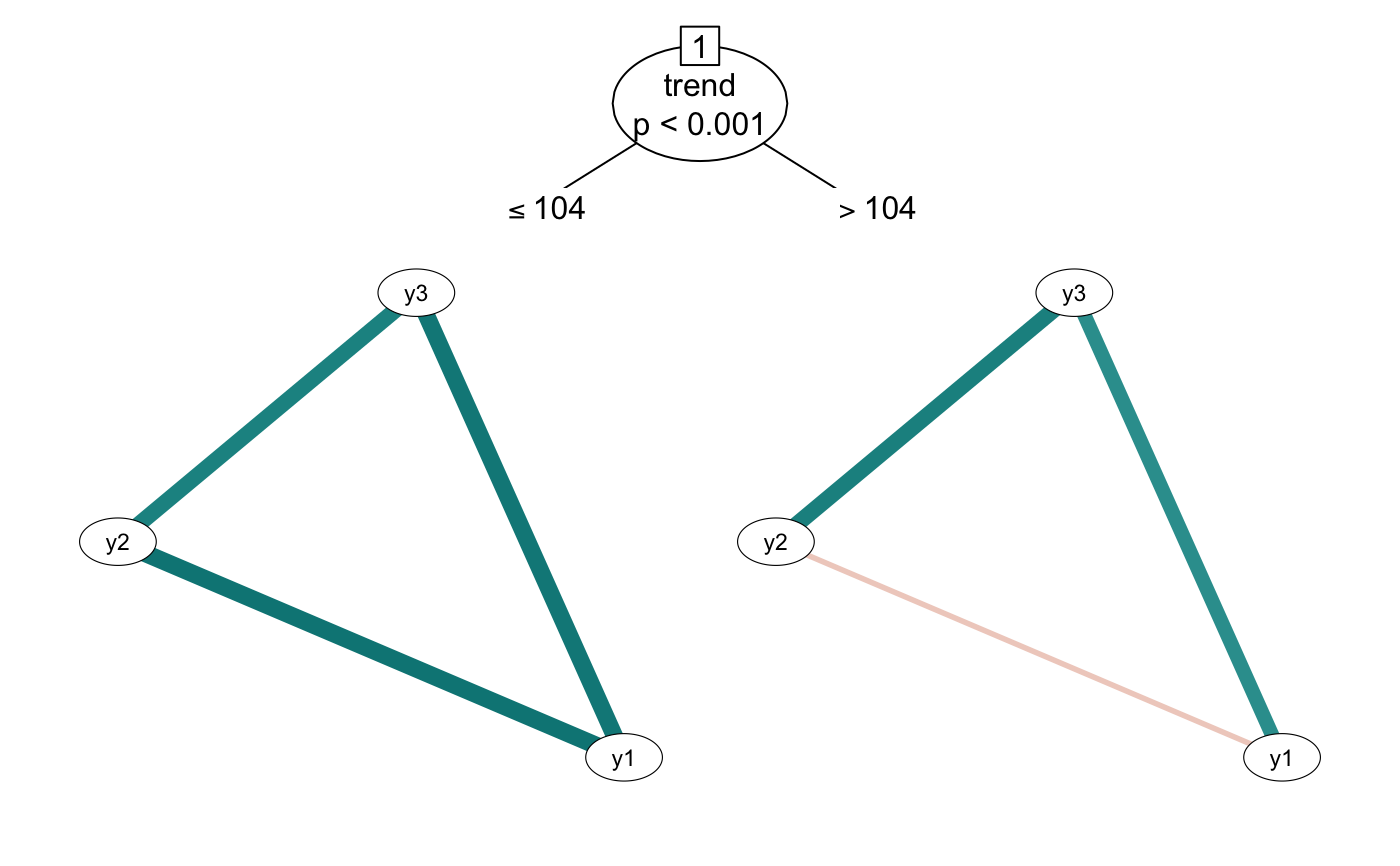
## plot
plot(tree2)
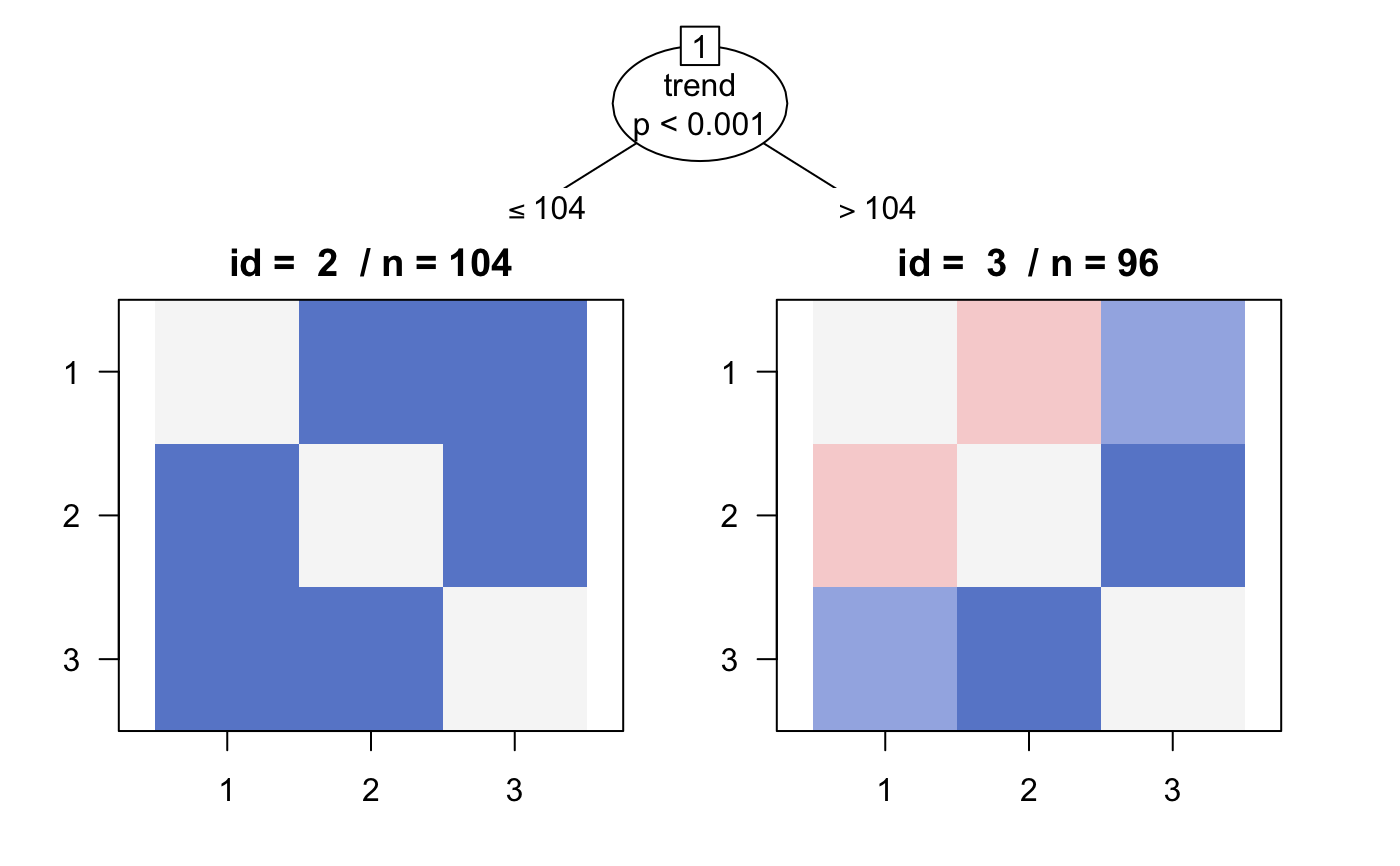
 plot(tree2, terminal_panel = "box")
plot(tree2, terminal_panel = "matrix")
# \donttest{
## Conditional version
tree3 <- networktree(nodevars=d[,3:5], splitvars=d[,1:2],
method="ctree")
## Change control arguments
tree4 <- networktree(nodevars=d[,3:5], splitvars=d[,1:2],
alpha=0.01)
# }
plot(tree2, terminal_panel = "box")
plot(tree2, terminal_panel = "matrix")
# \donttest{
## Conditional version
tree3 <- networktree(nodevars=d[,3:5], splitvars=d[,1:2],
method="ctree")
## Change control arguments
tree4 <- networktree(nodevars=d[,3:5], splitvars=d[,1:2],
alpha=0.01)
# }